Research
An overarching theme of my research is to characterize key health-related functions of the gut microbiome. I do this through examining gut microbial enzymes using comparative genomics and by developing computational tools.
Gut microbial enzymes
Gut microbes directly influence human health and disease by metabolizing diverse dietary and other host-derived compounds. Gut microbial enzymes are implicated in these functions by catalyzing key metabolic reactions, but most of them are yet to be identified. I use comparative genomics to identify gut microbial enzymes and characterize their evolutionary history, in addition to their taxonomic distribution. I collaborate with my wet lab colleagues to experimentally verify my predictions.
Relevant papers
- Zhou, Z., Jiang, A.K., Jiang, X. & Hatzios, S. K. Metabolic cross-feeding of a dietary antioxidant enhances anaerobic energy metabolism by human gut bacteria. Cell Host & Microbe S193131282500280X (2025) doi:10.1016/j.chom.2025.07.008.
- Arp, G., Jiang, A.K., Dufault-Thompson, K., Levy, S., Zhong, A., Wassan, J.T., Grant, M.R., Hall, B., Jiang, X.F.. Identification of gut bacteria reductases that biotransform steroid hormones. Nature Communications 16, 6285 (2025). https://doi.org/10.1038/s41467-025-61425-6
- Minabou Ndjite, G.†, Jiang, A.K.†, Ravel, C.T., Grant, M.R., Jiang, X.F., Hall, B. Gut microbial utilization of the alternative sweetener, D-allulose, via AlsE. Communications Biology 8, 970 (2025). https://doi.org/10.1038/s42003-025-08391-3
- Levy, S., Jiang, A.K., Grant, M.R. et al. Convergent evolution of oxidized sugar metabolism in commensal and pathogenic microbes in the inflamed gut. Nature Communications 16, 1121 (2025). https://doi.org/10.1038/s41467-025-56332-9 Code
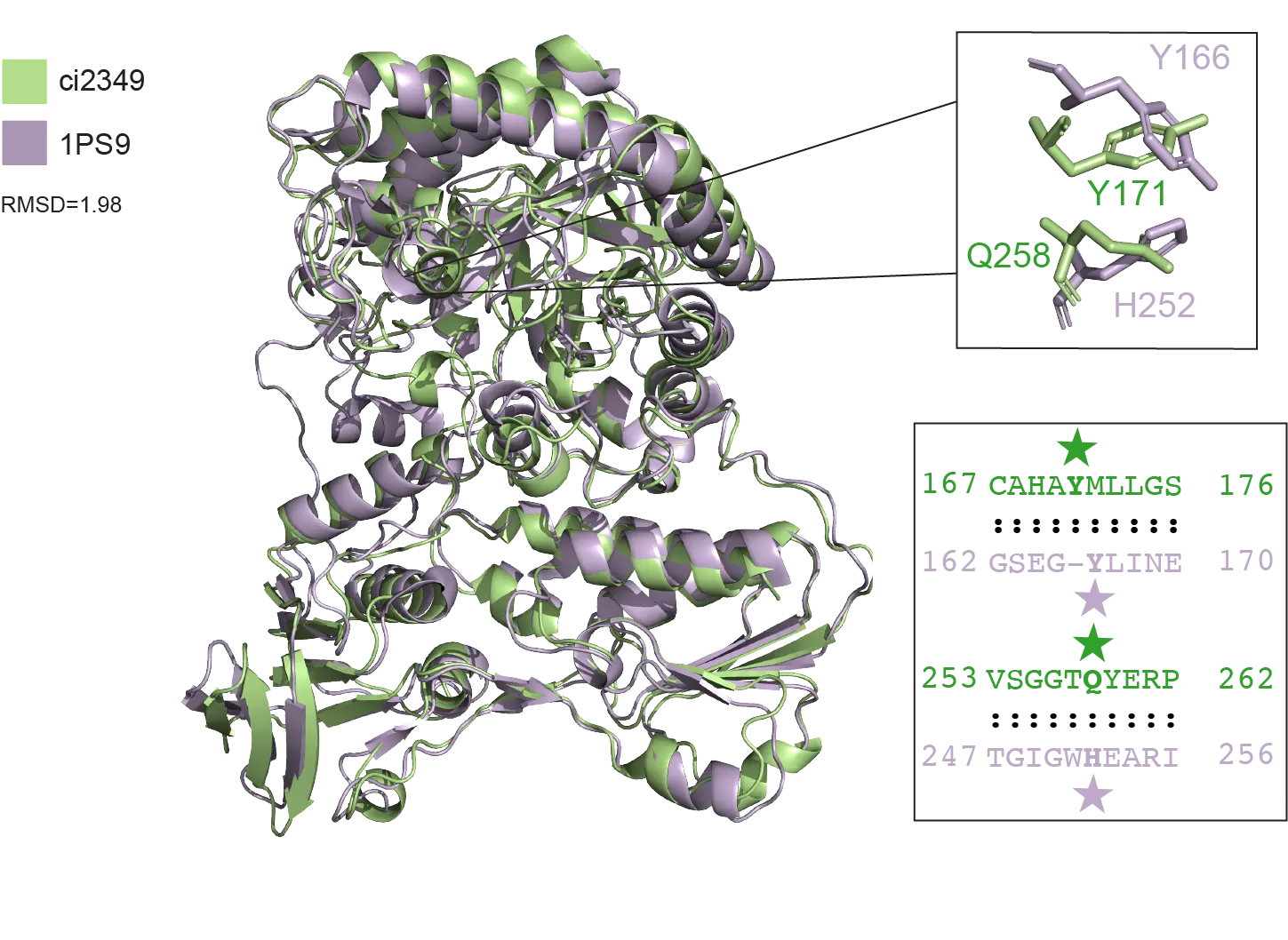
Comparison of a novel progesterone derivative reductase to E. coli 2,4-dienoyl CoA reductase
Computational genomics
I am interested in developing user friendly computational tools and pipelines to analze and interpret complex biological genomics data, particularly in relation to enzymes. I developed EzSEA (Enzyme Sequence Evolution Analysis), a web tool to identify evolutionarily important mutations in an enzyme using ancestral state reconstruction. You can check it out here.
Relevant papers
- Jiang, A.K.†, Zhao, J†, Jiang, X.F., EzSEA: An Interactive Web Interface for Enzyme Sequence Evolution Analysis, Bioinformatics Advances, vbaf118, (2025). https://doi.org/10.1093/bioadv/vbaf118

EzSEA web server homepage
